Twist of Two: 2D-LC for Oligos
💡Why Use 2D-LC for Oligo Analysis?
In oligonucleotide analysis, precision isn't just important, it's critical. However, analyzing them is no easy task. Oligonucleotides are complex molecules that often involve truncated sequences, structural variants, and impurities that pose challenges for conventional one-dimension separation methods. This is where two-dimensional liquid chromatography (2D-LC) comes in, it can improve the resolution, sensitivity, and reliability needed to truly see what is contained in each oligo sample.
💡 How Does 2D-LC Go Deeper Than 1D-LC?
For decades, one-dimensional liquid chromatography (1D-LC) has been the method of choice for biopharmaceutical characterization. But here’s the catch: its chromatographic selectivity is limited, its retention mechanisms lack orthogonality, and its peak resolution often falls short leading to poor separation. To break it down, when complexity rises, 1D-LC struggles to keep up. In best case, two-dimensional LC takes a separation to the next level by coupling two similar or different chromatographic modes, each targeting distinct molecular properties. So, the aim is to resolve coeluting compounds that 1D-LC can’t untangle (see figure 1).
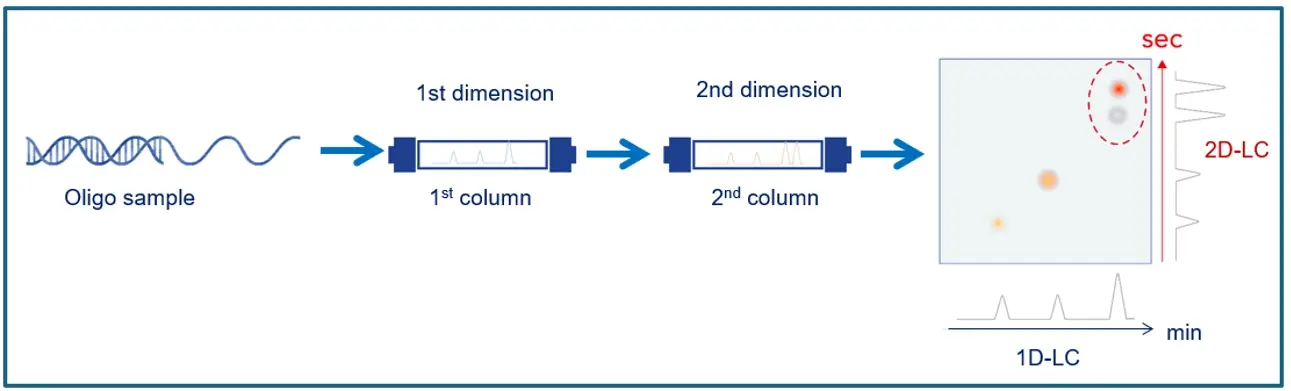
Figure 1: 2D-LC Principle (Graphic by KNAUER).
As a rule of thumb for the total analysis time, the second dimension sets the pace. Why? Because the slowest component in one run must finish before the fastest component of the next can even start. To be clear, analysis speed for second dimension separation isn’t just nice, it’s non-negotiable.
💡 What’s Inside a 2D-LC Setup?
At its core, 2D-LC is all about teamwork. Pumps and valves orchestrate the transfer between two chromatographic systems, each equipped with a column that uses similar or a different separation mechanism. Technically, 2D-LC uses two columns, two pumps, and smart valve switching, transferring fractions from the first to the second dimension in real time. Figure 2 illustrates example setup for comprehensive 2D-LC. It is highly recommendable that LC systems allow gradient elution in both dimensions, enabling highly efficient separations. The sample is introduced via an autosampler and first passes through the primary column (blue), where separation occurs. The eluent then moves with the peaks into the loop of a flow-switching valve or second autosampler unit. Once the loop is filled, the valve directs the collected separation window into the second-dimension column for further separation.
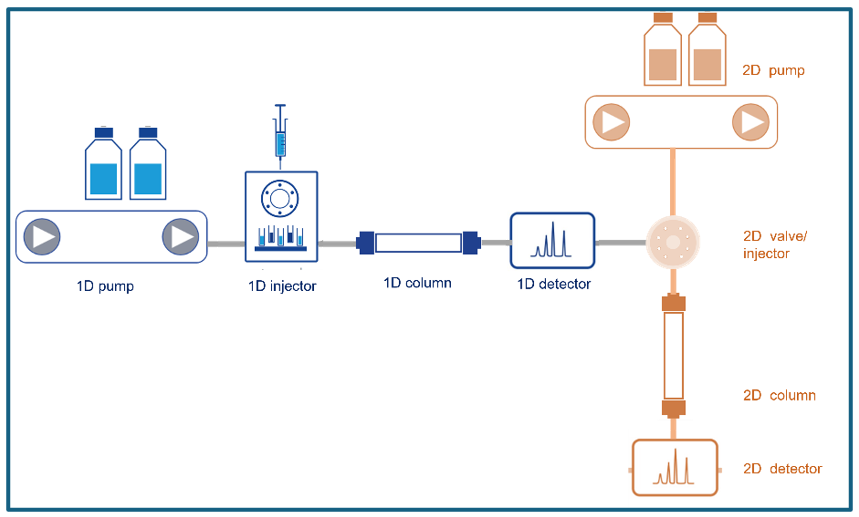 Figure 2:
2D LC System Configuration Example (Graphic by KNAUER).
Figure 2:
2D LC System Configuration Example (Graphic by KNAUER).
Think of the valve as the hidden hero of 2D-LC, because it’s the critical link that keeps both separation dimensions working in harmony and all under high pressure! The go to valve choices? Actually a ten-port or eight-port valve (two position) and different rotor settings can be used to realize a two-loop connection. You can also pair two six-port valves and sync their switching for seamless performance. In short, without the right valve, the magic of 2D-LC simply doesn’t happen. No matter which valves you choose, the principle stays the same. Once the analysis kicks off, the eluate from the first dimension fills one loop while the second-dimension pump flushes the other loop, sending its contents straight into the 2D column (see figure 3).
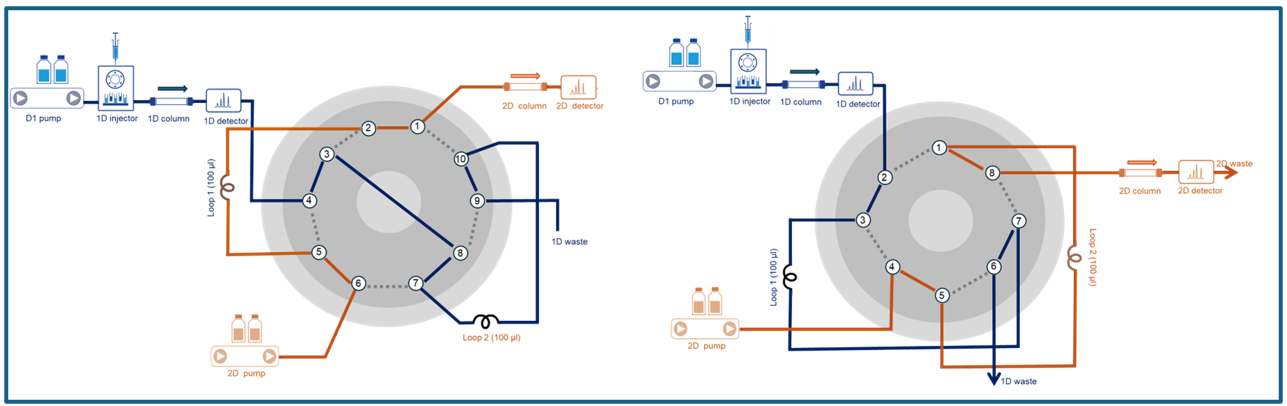
Figure 3: Schematic figure of ten-port (left) and eight-port valve (right) 2-position setup for 2D-LC (Graphic by KNAUER).
The KNAUER multifunction valve uses a cleverly arranged rotor setup (Figure 4) to realize this rhythmic dance! As one fills, the other empties. And the second-dimension separation must wrap up before the first loop finishes filling. So, timing is everything in 2D-LC!
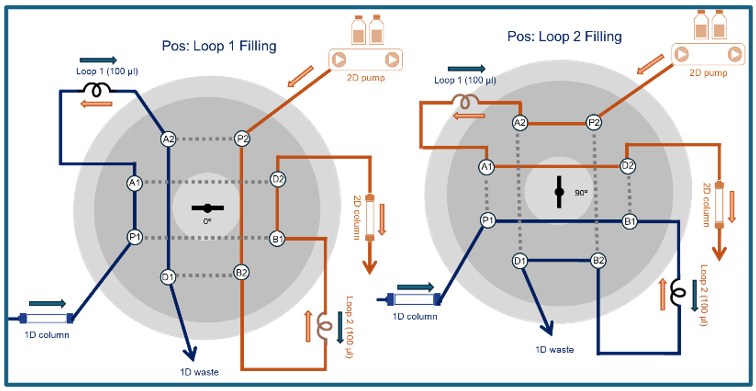
Figure 4: Schematic figure KNAUER eight-port 2 position valve for 2D-LC (Graphic by KNAUER).
💡 How Does 2D-LC Analysis Work for Oligos?
2D-LC pairs two separation columns in a single workflow. To tackle complex oligonucleotide samples the following combinations are possible:
1. 1st Dimension for Major Separation:
- Separation based on size (SEC), charge (AEX), or polarity (IP-RP, HILIC, Multi Mixed Chromatography - MMC), helping to reduce sample complexity.
- - Removes salts, buffers, and other matrix components that can interfere with downstream analysis.
- - Often applied as online desalting, ensuring MS-friendly conditions.
2. Peak transfer by Heart-Cutting (HC) or Comprehensive Transfer (CT):
- of interest are selectively transferred to the second dimension for deeper analysis with improved resolution and without column overload.
3. 2nd Dimension for High-Resolution Separation:
- Separation of closely related oligos and impurities, such as truncated sequences, chemical modifications, or sequence variants.
- The choice of mode (IP-RP, HILIC, AEX, or others) depends on the oligo type and analytical goal.
- Coupling with mass spectrometry allows precise identification and sequencing.
The magic lies in pairing two dimensions of different stationary phases (e.g., HILIC, IP-RP, AEX, MMC ect.) and/or applying different mobile phases on the same column type. This flexibility unlocks a variety of powerful separation workflows applied for Oligos:
- SEC x AEX [1]
- HILIC × IP-RP [2]
- IEC × IP-RP [3]
- IP-RP × IP-RP [4]
- AEX × HIC [5]
- MMC × desalting SEC [6]
- Chiral × RP [7]
💡 Full Picture or Just the Heart? Which 2D-LC Strategy Works for Your Oligos?
When it comes to 2D-LC for oligonucleotides, not all peaks are created equal. Heart-Cut (HC) and Comprehensive Transfer (CT) offer two very different approaches to navigating complex mixtures of oligos. What is the difference? If analyzing oligonucleotides, sometimes you don’t need the whole picture, you need the right slice. That’s where the heart cut 2D-LC approach shines. Instead of transferring every fraction from the first dimension, you select the most relevant peak region, the “heart” and send it to the second separation column for more resolution (see figure 5). The main advantage of this transfer approach is the second dimension isn’t in a hurry. You can adjust your analysis time, crank up the resolution, and separate even the trickiest peaks, exactly where you need the power to solve the oligo puzzle. Heart-cut 2D-LC is all about focus and efficiency, but it comes with a catch. By zooming in on just the selected peak region, you might miss unexpected impurities lurking outside the spotlight. That’s why it’s not the best choice for full characterization or completely unknown samples, it’s precision over total coverage.
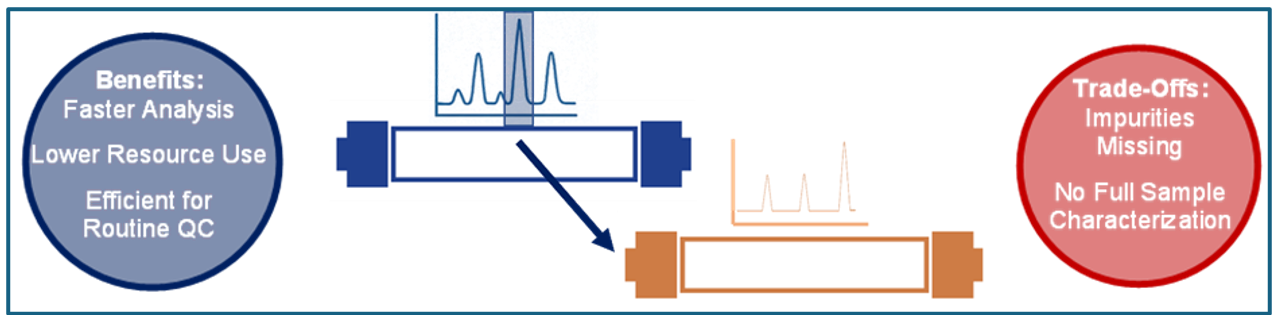
Figure 5: 2D-LC Single Heart-Cut Principle (Graphic by KNAUER).
Comprehensive transfer mode, on the other hand, goes all in (see figure 6). Every fraction from the first dimension is transferred to the second, giving you a complete, high-resolution picture of your sample’s complexity. Think of it as the full story, not just the highlights.
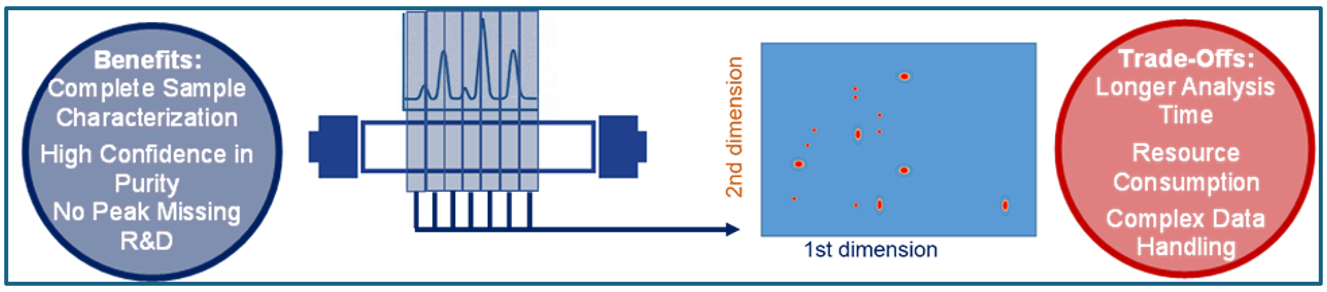
Figure 6: Full Comprehensive 2D-LC Principle (Graphic by KNAUER).
In a nutshell, comprehensive transfer captures every peak, making it the first choice for R&D, detailed impurity profiling, and regulatory submissions. It gives you the complete story of your oligonucleotide sample. Heart-Cut transfer has a focus where it counts. It concentrates on known critical peaks, making it perfect for routine QC or targeted analysis.
Sometimes, the best results come from blending strategies. The combination of both approaches is also sometimes used, we zoom in on the most relevant 1D section, packed with target analytes, and give it the full two-dimensional treatment.
💡 How to Master Oligo 2D-LC?
- Column Selection: Pair different or
same column types for
 orthogonal separation, the only limit is the mobile phase
compatibility. Always consider first dimension for resolution (--> Oligo size dominant), second dimension for MS-friendly conditions
(--> Oligo sequence dominant).
orthogonal separation, the only limit is the mobile phase
compatibility. Always consider first dimension for resolution (--> Oligo size dominant), second dimension for MS-friendly conditions
(--> Oligo sequence dominant).

Combine different pH values and organic modifiers for fine-tuned separation, apply also gradient mode in both dimensions to achieve maximum peak capacity. “Wrong” 1st Dimension mobile phase can destroy separation in 2nd Dimension. Design mobile phases as paired system. Use volatile salts (e.g., ammonium acetate) for MS-friendly workflows.
 3. Instrumentation I:
2D-LC Oligo analysis demands fast and robust valve design and switching,
precise timing, seamless fraction transfer, and absolute reproducibility. Optimize
the interface/valving between two columns (setup matters). Pressure limit of
the 2D switching valve must be considered in terms of 2D column back pressure.
3. Instrumentation I:
2D-LC Oligo analysis demands fast and robust valve design and switching,
precise timing, seamless fraction transfer, and absolute reproducibility. Optimize
the interface/valving between two columns (setup matters). Pressure limit of
the 2D switching valve must be considered in terms of 2D column back pressure.

4. Instrumentation II: Maximum transfer loop volume is defined by loading volume of 2D. Flow rate of first dimension is determined by the loop volume and the analysis time in 2D-LC. As a rule of thumb, loops should not be filled with more than 70% to avoid sample loss by parabolic flow profile.
 5. Method Development:
Start with scouting gradients in both dimensions, optimize for impurity
resolution. Separation in 2D must be as high as possible. Apply fast 2D
separations (à short columns, rapid gradients) to avoid peak
broadening and ensure sufficient sample frequency of 1D eluate. Adjust 2D cycle
time according to average width of 1D peak (should be smaller).
5. Method Development:
Start with scouting gradients in both dimensions, optimize for impurity
resolution. Separation in 2D must be as high as possible. Apply fast 2D
separations (à short columns, rapid gradients) to avoid peak
broadening and ensure sufficient sample frequency of 1D eluate. Adjust 2D cycle
time according to average width of 1D peak (should be smaller).
 6. Method
Optimization: First, optimize separation for second dimension. Focus on sampling
frequency (number of fractions per peak). To maintain resolution transfer timing is everything! It
is recommended to collect at least three, ideally four fractions while the peak
of interest is eluting in the first dimension. Final step is to optimize the
6. Method
Optimization: First, optimize separation for second dimension. Focus on sampling
frequency (number of fractions per peak). To maintain resolution transfer timing is everything! It
is recommended to collect at least three, ideally four fractions while the peak
of interest is eluting in the first dimension. Final step is to optimize the
 separation of the first dimension, including flow rate, analysis time and
gradient.
separation of the first dimension, including flow rate, analysis time and
gradient.
💡 Final Thoughts
2D-LC pairs distinct separation techniques to tackle even the most complex mixtures, think oligos with peak coelution. By combining orthogonal separation mechanism, it reveals hidden details and sharper insights that 1D-LC simply can’t deliver. Unlocking this power isn’t plug-and-play. True orthogonality must be achieved and measured, mobile phases need to play nicely across dimensions, and sampling must be fast enough to prevent peaks from remixing. In short, mastering 2D-LC is a balancing act of column chemistry, timing, and precision.
If you're planning to set up or optimize your oligonucleotide workflow, feel free to contact us at sales@knauer.net. Stay tuned for more exciting insights into the Oligonucleotide world in our “Oligos Made Easy” series.
For more in-depth discussion or questions, reach out to the author at kindler@knauer.net
Literature:
F. Hannauer, R. Black, A.D. ray, E. Stulz, G.J. Langley, S.W. Holman, “Advancements in the characterization of oligonucleotides by high performance liquid chromatography-mass spectrometry in 2021: A short review,” Anal Sci Adv., 3. 90–102 (2022)
[1] Sartorius BIA Separations (Poster): 2D-LC SEC-AEX Evaluation of Complex Samples Containing Extracellular Vesicles (2024)
[2] Q. Li, F. Lynen, J. Wang, H. Li, G. Xu, and P. Sandra, “Comprehensive Hydrophilic Interaction and Ion-Pair Reversed-Phase Liquid Chromatography for Analysis of Di- to Deca-Oligonucleotides,” Journal of Chromatography A 1255, 237–243 (2012)
[3] S. G. Roussis, I. Cedillo, and C. Rentel, “Two-Dimensional Liquid Chromatography-Mass Spectrometry for the Characterization of Modified Oligonucleotide Impurities,” Analytical Biochemistry 556, 45–52 (2018)
[4] D. Stoll, M. Sylvester, D. Meston, M. Sorensen, and T. D. Maloney, “Development of Multiple Heartcutting Two-Dimensional Liquid Chromatography With Ion-Pairing Reversed-Phase Separations in Both Dimensions for Analysis of Impurities in Therapeutic Oligonucleotides,” Journal of Chromatography A 1714, 464574 (2024)
[5] A. Kazarian, J. Lee, B. Mitasev, and H. Choi, “Mixed-Mode Separation of Antisense Oligonucleotides Using a Single Column With Complementary Anion-Exchange and Hydrophobic Interaction Chromatography Approaches,” Journal of Chromatography A 1740, 465581 (2025)
[6] F. Li, X. Su, S. Bäurer, and M. Lämmerhofer, “Multiple Heart-Cutting Mixed-Mode Chromatography-Reversed-Phase 2D-Liquid Chromatography Method for Separation and Mass Spectrometric Characterization of Synthetic Oligonucleotides,” Journal of Chromatography A 1625, 461338 (2020)
[7] F. Li, C. Knappe, N. Carstensen, et al., “Two-Dimensional Sequential Selective Comprehensive Chiral×Reversed-Phase Liquid Chromatography of Synthetic Phosphorothioate Oligonucleotide Diastereomers,” Journal of Chromatography A 1730, 465076 (2024)

