
Science with Passion
Application No.: VPH0076 Version 1 10/2022
Development and optimization of LNP formulations – using the KNAUER NanoScaler
Josep Garcia, Carles Felip; Curapath, Av. Benjamin Franklin 19 46980 Paterna, Valencia (Spain), www.curapath.com
J. Wesolowski, Y. Krauke, G. Greco; krauke@knauer.net
KNAUER Wissenschaftliche Geräte GmbH, Hegauer Weg 38, 14163 Berlin
Rendering: Curapath
Summary
Lipid nanoparticles (LNPs) offer significant advantages as a delivery system for nucleic-acids therapeutics. In addition to functional advantages, such as biocompatibility, biodegradability, and high entrapment efficiency, LNPs can be manufactured rapidly, reproducibly, and with high scalability. At Curapath, we optimize LNP formulation during pre-clinical research and development in order to determine the optimal conditions for encapsulation of nucleic acids. This application note describes the use of the NanoScaler, KNAUER’s new Impingement Jets Mixing (IJM) benchtop system, to formulate LNPs of adequate size and PdI, that are optimally able to encapsulate various nucleic-acid payloads with high reproducibility and efficiency. The pandemic proved the IJM to be a vertically integrated LNP-production platform allowing for seamless transfer from alternative mixing technologies, and a like-for-like technology to be used at every stage of the scale-up process. From preclinical to commercial, we have a path forward.
Introduction
The unprecedented success of the SARS-CoV-2 vaccines BNT162b2 (“Comirnaty” from BioNTech/Pfizer) [1] and mRNA-1273 (“Spikevax” from Moderna) [2], has prompted pharmaceutical companies to focus on the use of lipid nanoparticles (LNPs) for the delivery of nucleic acids. Examples abound of the use of these non-viral vectors in ongoing clinical trials aimed at the prevention and treatment of infections, cancer, and genetic diseases. The manufacture of LNPs is a complex process that must be robust, scalable, and reproducible, in which the Impingement Jets Mixer plays a critical role (Fig. 1). To meet such needs, KNAUER has designed an Impingement Jets Mixing (IJM) technology, enabling precise mixing of organic phase containing the lipids and the aqueous phase containing the nucleic acids. Tightly controlling the Jets mixer operating parameters allows the system to produce LNPs of reproducibly defined size. KNAUER’s IJM technology has been proven in the formulation of LNPs for large-scale manufacturing of SARS-CoV-2 vaccines. KNAUER’s new benchtop IJM system, the NanoScaler (Fig. 2), is designed for low sample consumption, thus facilitating the optimization of process parameters. Using the NanoScaler at Curapath, we assessed the effect of varying key operating parameters on LNP size, Pdl, and encapsulation efficiency. Additionally, we evaluated the effect of total flow rate, and formulated a 50 ml exemplary LNP drug product. The information emanating from this work serves as practical guidance for the development and optimization of LNP formulation and demonstrates the versatility of the NanoScaler system to produce from 1 ml to various liters of LNP formulations within the same mixing technology.

Fig. 1 Process workflow for LNP formulation using NanoScaler.

Fig. 2 The NanoScaler and its components
Results
We used the NanoScaler to formulate LNPs with mRNA (CleanCap Fluc, 5moU, TriLink) and pDNA (pCMV-Luc, PlasmidFactory) payloads. The first approved LNPs formulation (ONPATTRO®) was used as reference, with minor modifications as described in Tab. 1.
Tab. 1 LNPs composition used in all the study

As initial parameters, the total flow rate (TFR) was set to 2 ml/min and the ratio between aqueous phase and organic phase to 3:1. The aqueous phase containing nucleic acids was injected via a 1 ml loop designed to minimize waste volume. We used the smallest IJM (IJM 1) for low TFR. Following formulation, LNPs were diluted in PBS at pH 7.4 and concentrated by centrifugation with the Vivaspin™ 500 (Sigma) to remove ethanol. All experiments have been performed in triplicate.
The size of the thus formulated pDNA-LNPs was larger than that of the mRNA-LNPs, ranging from 110 to 130 nm and from 80 to 100 nm, respectively (Fig. 3). This was expected, given that the encapsulated pDNA is much larger, at 6.2 kbp, and bulkier than the mRNA counterpart, at just 1.9 kb. The PdI of both formulations was quite adequate at less than 0.2.

Fig. 3 Size and PdI of LNP formulations containing different nucleic-acid payloads
The NanoScaler was then run alongside the Herringbone (Darwin Microfluidics), a system having a traditional microfluidics-chip pattern, to formulate pDNA- and mRNA-LNPs.
In general, the IJM-based NanoScaler and the Herringbone produced LNPs of similar characteristics (Fig. 4). The Pdl of all four groups of LNPs was low, with those formulated by the NanoScaler showing somewhat higher reproducibility. The two systems yielded pDNA-LNPs of nearly identical size, which were larger than the mRNA-LNPs. Interestingly, mRNA-LNPs formulated by the NanoScaler were smaller than mRNA-LNPs from the Herringbone.
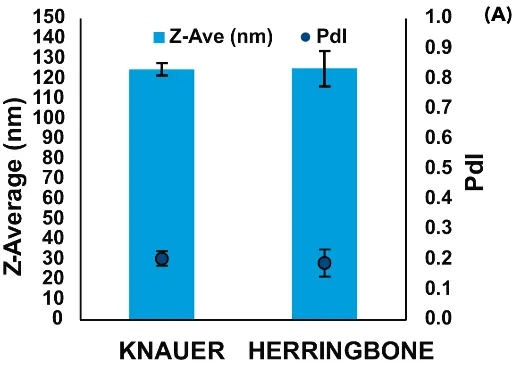
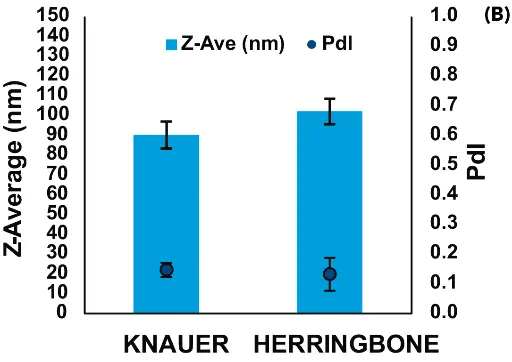
Fig. 4 Comparison of LNPs formulated with the NanoScaler and Herringbone micromixers. A) pDNA-LNPs, and B) mRNA-LNPs.
One of the most important parameters to optimize when formulating LNPs is the TFR. In order to test the effect of flow rate on LNP size, Pdl, and encapsulation efficiency, we ran the NanoScaler with mixer 1 at TFRs ranging from 1 to 5 ml/min, keeping all other parameters unchanged.
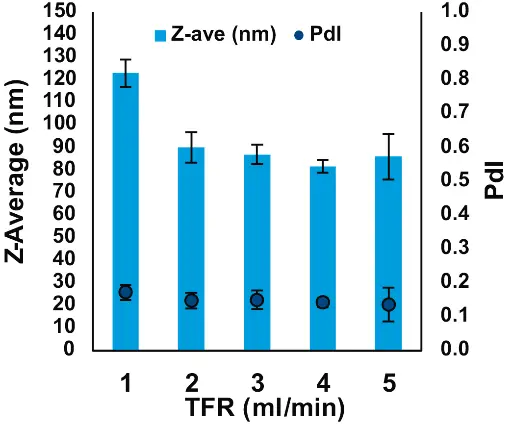
Fig. 5 Effect of TFR on size and PdI of mRNA-LNPs formulated with the NanoScaler
We found that increasing TFR from 1 to 2 ml/min sharply reduced the size of the mRNA-LNPs (Fig. 5). A slight trend of decreasing LNP size was observed when TFR was increased from 2 to 3 and 4 ml/min. This trend was broken at 5 ml/min, a flow rate that also showed greater variability of both the size and Pdl of the LNPs. Based on these results, we would recommend setting the TFR at between 2 and 4 ml/min, if the desired LNP size is from 70 to 90 nm, as is the case for ONPATTRO®. Encapsulation efficiency (EE%) was measured using the Quant-it RiboGreen™ assay. The EE% of the mRNA-LNPs was greater than 90 % across all TFR tested (Fig. 6).
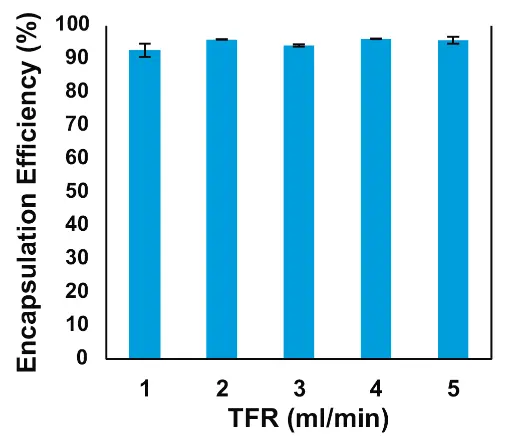
Fig. 6 Effect of TFR on encapsulation efficiency of mRNA-LNPs formulated with the NanoScaler
The initial concentration of lipid and nucleic acids can be adjusted to meet client needs. Higher initial concentrations can be used to shorten the production time of large-scale batches, whereas lower concentrations are used in R&D runs in order to decrease the amounts of lipids/nucleic acids used.
We tested the effect of varying the initial lipid and nucleic-acid concentration on LNP properties. Other operating parameters were kept as described above, except that TFR was set at 3 ml/min.


Fig. 7 Effect of initial lipid and nucleic-acid concentration on mRNA-LNPs (A) and pDNA-LNPs (B) formulated with the NanoScaler
We observed no significant difference in size and PdI among all the pDNA-LNPs formulated (Fig. 7). However, at the concentration of 1.4 mg/ml, the size of mRNA-LNPs was significantly larger, and Pdl showed much greater variability. Unlike the 1.4 mg/ml concentration, the larger concentrations tested proved to be adequate for formulating mRNA-LNPs.
In-line dilution of LNPs is a process improvement that allows greater automation and facilitates scale-up. We Incorporated in-line dilution into the NanoScaler process, setting TFR at 3 ml/min and flow through the dilution pump at 1.5 ml/min, thus keeping the dilution flow at the usual 50 % of TFR. We observed that the mRNA-LNPs obtained with an in-line dilution were not significantly different from undiluted LNPs (Fig. 8), thus demonstrating the scalability of the NanoScaler system for manufacturing larger batches of LNPs.
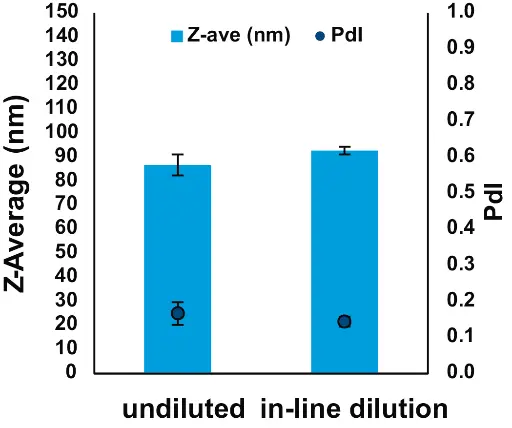
Fig. 8 Effect of in-line dilution on mRNA-LNPs formulation with the NanoScaler
We were also able to formulate batches 50 times larger than R&D batches, obtaining mRNA-LNPs of similarly adequate characteristics (Fig. 9). Preclinical studies can therefore be performed with the use of the NanoScaler, as several hundred mL of LNPs can be produced in a few hours.
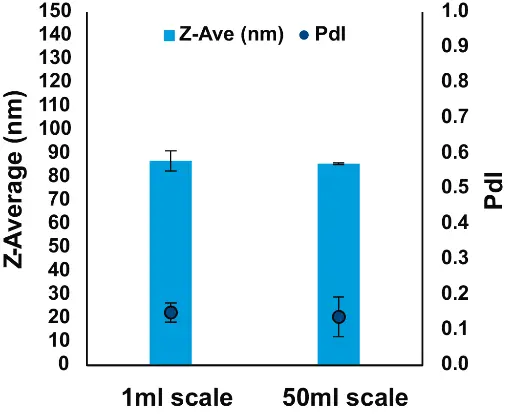
Fig. 9 Effect of scale-up on size and PdI of mRNA-LNPs formulated with the NanoScaler
At Curapath, a 50 ml batch is formu-lated in less than 20 min, obtaining similar results as with 1-ml R&D batches. The larger batches are then purified by TFF hollow fiber 20 cm PES 50KDa MWCO columns, filtered through optimized membrane Sartopore® Platinum Sartoscale 0.2 µm filters, and poured into glass vials, simulating the filtration sterilization procedure. (Fig. 10 and Tab. 2).

Fig. 10 Finished vial containing mRNA-LNPs from large-scale batch
Tab. 2 Values obtained for IPCs and final QC in the exemplary formulation following workflow process shown in Fig. 1
IPC | Size (nm) | PdI | mRNA conc (ng/µL) | EE (%) |
1 | 62 | 0.28 | 69.4 | 95.5 |
2 | 88 | 0.18 | 28.2 | 97.0 |
3 | 94 | 0.21 | 218.6 | 94.5 |
Final QC | 86 | 0.14 | 223.2 | 96.5 |
To scale-up the process, the NanoScaler offers five different mixer sizes, allowing operations in a wide TFR range. In the next example, mixer number 2 was tested increasing the TFR from 4 to 10 ml/min (Fig. 11).

Fig. 11 Comparison of size and PdI of mRNA-LNPs formulated with NanoScaler IJM mixer 2 at various TFR rates
The LNPs formulated with IJM 2 (Fig. 11) showed characteristics similar to LNPs obtained with IJM 2 (Fig. 5), further confirming that the NanoScaler can be successfully used to formulate large-scale batches, and is well suited for the manufacturing environment. A complete and efficient cleaning of the system can be achieved in less than 3 hours by flushing at 20 % of the maximum flow rate of the pump head with H2O, EtOH and 0.1 M NaOH. After the cleaning protocol, a sample was collected from the instrument and analyzed for nucleic acid/lipids detection (Tab. 3).
Tab. 3 Results obtained to prove the presence of lipid or API impurities
Compound | Instrument | Amount detected |
mRNA/pDNA | Stunner | ND |
MC3 | HPLC-CAD | ND |
HPLC-CAD | ND | |
HPLC-CAD | ND | |
HPLC-CAD | ND |
Lastly, we assessed the potency of nucleic-acid-LNPs by in vitro transfection into various cell lines. Cells were seeded into 96 well plate at: HepG2: 30,000 cells/well; HEK 293:15,000 cells/well; HeLa: 8,000 cells/well. After 24h, the LNPs were added at 300 ng of mRNA/pDNA per well. Luminescence was measured after incubation for 24 hours.

Fig. 12 Luminescence of cells incubated with mRNA- and pDNA-LNPs relative to non-treated cells. compared to non-treated cells
Luminescence data clearly demonstrates that LNPs containing either mRNA or pDNA enter cultured cells of the Hep G2, HeLa, and HEK cell lines upon incubation. mRNA-LNPs showed far greater potency (Fig. 12). This is to be expected because LNP formulation has been optimized for mRNA delivery, and because they are considerably smaller than pDNA-LNPs. Also noteworthy is that Hep G2 cells produced many times more luminescence than HeLa and HEK cells. Being derived from liver, Hep G2 cells have a higher rate of internalization and are more easily transfected.
Conclusion
KNAUER’s new IJM system, the NanoScaler, is able to encapsulate mRNA and pDNA into LNPs with high reproducibility and efficiency. At Curapath, the performance of the NanoScaler for formulating LNPs matched that of the traditional Herringbone. In addition, the NanoScaler offers flexibility and control that enable the user to tune the size of LNPs by increasing or decreasing the TFR. The properties of LNPs formulated with the NanoScaler are not affected by the initial lipid/nucleic-acid concentration, making it possible to work with lower initial concentrations in R&D, and higher concentrations when manufacturing large-scale batches in order to shorten production time. The use of the NanoScaler for large-scale production of LNPs is also facilitated by the availability of variously sized IJM mixers. The versatility of the equipment permits the client to scale up the LNP production by using the different IJM mixers. The smaller IJM mixer (number 1) suits perfectly for R&D purposes and, by using the IJM number 5, the equipment is able to produce a few liters of LNPs in less than 2 hours. It is also possible to incorporate in-line dilution to lower the ethanol concentration prior to purification without affecting LNP characteristics. Notably, after formulation, all the parts of the equipment can be easily cleaned with basic solvents such as water, ethanol and sodium hydroxide; removing all the LNPs components from the previous production. Very importantly, we have shown that the LNPs formulated with the NanoScaler are able to transfect various commonly used cell lines with mRNA and pDNA payloads.
Materials and Methods
The IJM NanoScaler (Knauer) consisted of 5 impingement jets mixers (IJM), 3 pumps and 2 Valves. Final method for small scale LNPs production was as follows: Number 1 IJM was used with TFR ranged from 1–5 ml/min and aqueous:organic phase ratio 3:1. Initial lipid concentration ranged from 1.4–11.4 mg/ml. LNPs were diluted after formulation in PBS 7.4 and VivaspinTM 500 (Sigma) was used to remove ethanol. For large scale LNPs the same conditions were followed but Number 2 IJM was used with TFR set in the range of 4–10 ml/min and TFF hollow fiber 20 cm PES 50KDa MWCO was applied instead of VivaspinTM to remove ethanol. Payload encapsulation efficiency was obtained using the Quant-it
RiboGreenTM (for mRNA or pDNA) assays in all cases. Size and polydispersity were determined by dynamic light scattering (Stunner, Unchained labs). For a head-to-head comparison with IJM NanoScaler the conventional microfluidics device with Herringbone pattern (Darwin MicroFluidics) was used. Transfection efficiency in HepG2 cells, HEK 293 cells and HeLa cells was determined by luminescence (Bright-Glo™ Luciferase Assay, Promega) upon 24 hours of LNPs incubation.
References
[1] Anderson, E. J., Rouphael, N. G., Widge, A. T., Jackson, L. A., Roberts, P. C., Makhene, M., & Beigel, J. H. (2020). Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. New England Journal of Medicine, 383(25), 2427-2438.
[2] Polack, F. P., Thomas, S. J., Kitchin, N., Absalon, J., Gurtman, A., Lockhart, S., & Gruber, W. C. (2020). Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. New England Journal of Medicine.
[3] Hou, X., Zaks, T., Langer, R., & Dong, Y. (2021). Lipid nanoparticles for mRNA delivery. Nature Reviews Materials, 6(12), 1078-1094.
[5] https://investors.biontech.de/covid-19-vaccine-partners
[6] https://edition.cnn.com/2021/03/31/health/pfizer-vaccine-manufacturing/index.html
Application details
Method | LNP (Encapsulation) |
Mode | IJM (Impingement jets mixing vs. microfluidic herringbone mixing) |
Substances | mRNA, pDNA |
Version | Application No.: VPH0076 Version 1 | 10/2022 | ©KNAUER Wissenschaftliche Geräte GmbH |
